Prev: Novel MRI data analysis algorithms and tools Next: Publications
Project Results: Clinical Decision Support by Quantitative MRI
The first project goal was to develop a retrospective multi-parametric classification and clustering approach on a group level based on features extracted from previously acquired multi-modal quantitative MRI data sets and machine learning. For the development and testing of different clustering and classification algorithms pre-existing patient data acquired with consistent scan protocols from cohorts of patients with major depressive disorder (MDD) and multiple sclerosis (MS) were used. All data were consistently processed and prepared for classification trials. Subsequently, the implementation of different machine learning methods such as support vector machines (SVM), random forest (RF), k-Nearest Neighbors and multiple kernel learning (MKL) approaches were performed to investigate their performance in different prediction tasks. Early stage efforts demonstrated that distinguishing the patient groups from healthy volunteers was possible for both diseases, MDD and MS. In a second phase more challenging tasks were studied (differential diagnostics; treatment responders versus non-responders; patients with different clinical subtypes; subjects with different disease progression) and provided good specificity and sensitivity. Finally, the focus shifted onto prospective classification of single patient data sets by refining the classification and clustering methods for the more difficult prediction tasks and by optimizing the combination and selection of multi-modal quantitative MRIreadouts. The classification module was integrated into the software framework and applied as a prospective clinical decision support system for MDD and MS based on the most successful classifiers using data acquired both prior and during the project run time for multiple sclerosis and major depressive disorders. It was demonstrated that anatomical, functional and metabolic MRI yield features that predict therapy response to electroconvulsive, Ketamine therapy and psychotherapy in patients with major depressive disorder. Furthermore, the correct assignment of patients to different types of multiple scleroses with distinct disease progression was possible based on a combination of anatomical and microstructural MRI. Microstructural MRI shows strong predictive power for disability scores in multiple sclerosis patients. In both patient groups it was found that anatomical imaging data that are routinely acquired yield predictive power if a quantitative analysis is performed instead of the qualitative evaluation that is the current clinical standard.
Demonstration of Clinical Decision Support in MDD
Next to diagnostic classification tools, classifications routines for the prediction of treatment responses have been developed. Pre-existing datasets of MDD patients that received Electroconvulsive Therapy (ECT), or ketamine treatment have been processed. The next paragraphs summarize the most important findings.
The Major Depression Study includes multiple samples of patients affected by Major Depressive Disorder (MDD) and Healthy Controls (HC) subjects, acquired in different modalities (Structural, resting-state and task-based fMRI), at different time points (before and after treatment) and at different sites (Berlin and Zurich). The main focus was the development of an optimal and interpretable prediction pipeline, that is applicable for the diverse set of input data and clinical tasks. The goal was to obtain a robust and accurate predictive model, providing interpretable results while identifying the informative modalities for a specific clinical task.
Prospective Clinical Trial in Major Depressive Disorder
Prospective multimodal QuaMRI dataset including MDD patients and healthy control participants were acquired at one partner site. Acquisition sequences included a 2D J-semiLASER magnetic resonance spectroscopy (MRS) sequence, an arterial spin labeling (ASL) sequence and a diffusion tensor imaging (DTI), all developed in the CDS-QuaMRI project. The multimodal MRI data was acquired at multiple time points (baseline, 24 h post treatment, 4 weeks post treatment, 6 months post treatment), to develop classification routines that take into account early brain changes during different antidepressant treatments and to make predictions on long-term outcomes based on these early changes. Long-term follow-up measurements will be conducted beyond the project end. The status at the end of the project duration is given in Table 1.
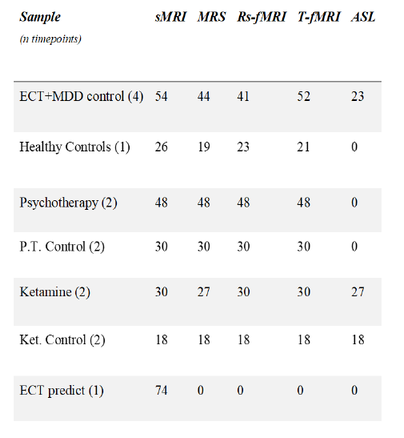
Table 1: Final status of the data acquisition. The numbers depict the N that has been acquired for each data set and modality.
Clinical task: patients vs controls
In the first phase of the project, focus lay on the development of the best classification approach, with the aim of distinguishing patients and controls. The disease diagnosis task was based on a dataset of 118 samples of task-based fMRI data. This effort resulted in the development of a multi-voxel pattern classification pipeline. The machine learning analysis employed feature selection and linear Support Vector Machines to distinguish between healthy controls and depressed patients based on whole-brain fMRI data. The post processing cluster analysis identified brain regions that mostly contributed to the classification, i.e. activation patterns in patients or controls. The results of this approach showed good classification performance, with accuracy reaching 71%. Details on the analysis and discussion of the corresponding findings can be found in the published manuscript.
Subsequently, the focus shifted to studying whether a multi-modal approach is superior over a single-modality study. For this task, resting-state and Structural MRI data from the same cohort of MDD and HC subjects was used. The results of Multiple Kernel Learning (MKL) techniques and feature concatenation strategies were compared. It was observed that a combination of contrast images obtained from task-based fMRI provide the best results with linear classification methods. When non-linear kernels are used, the single best performing modality dominates over the multi-modal counterparts. With the same method, it has also been observed that combining resting state and task-based fMRI improves over the individual modalities. Overall, there was no clear benefit in using complex MKL techniques over simple feature concatenation strategies. In general, the improvements observed with modality combinations are not consistent across methods and modalities. The outcome is certainly influenced by the limitations given by a small sample size. This analysis suggests that combining multiple modalities is promising and has the potential to improve the automated clinical decision system. Still, more data should be acquired in order to provide a final complete assessment. Enlarging the sample size would also be beneficial for model exploration, allowing the use of deep learning based strategies or transfer learning techniques.
Clinical task: response prediction
In personalized medicine, a highly relevant task is the identification of patients that positively respond to a given treatment based on their clinical characteristics. In the final stage of the CDS-QuaMRI project the aim was to assess response prediction to Electroconvulsive Therapy (ECT) treatment using MRI data. The question was investigated both from a classification and regression perspectives, while structural MRI features were used for the analysis. Initially, a whole-brain approach based on Support Vector Machines was developed to classify responders versus non responders. A good predictive performance was obtained with 26/39 responders and 23/32 non-responders being correctly identified. The post processing analysis showed a cluster in the right anterior parahippocampal gyrus (aPHCr) region which provided the most informative contribution in the characterization of ECT response. Then, in the second step the same region was used within a regression model to predict the percentage of symptom reduction (PSR). The results showed a significant correlation between predicted and true PSR, indicating that the aPHCr area contains the relevant information to identify responders (Figure 1). The results of this pioneer study confirm that there is predictive power in structural brain images of MDD patients to predict ECT response and that this method can be used to predict the treatment response in single patients. Future work should focus on integrating the multi-modal analysis applied for the patients versus control task with the ECT classification task, and extend it to the regression problem.
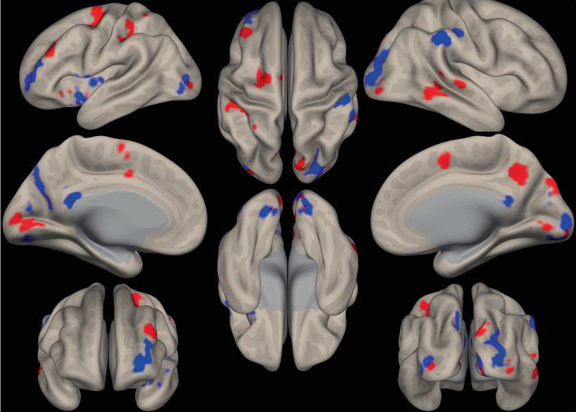
Fig. 1 SVM weight map. The location of the most relevant SVM classification weights from the WM > fixation contrast are shown (20% of the highest weights with a cluster threshold of 50 voxels). Red regions depict more activation in MDD patients. Blue regions depict more activation in healthy controls.
Demonstration of prospective Clinical Decision Support in MS
Multiple Sclerosis is a neurological disease of the central nervous system, whose cause and progression is still unknown. Patients can be diagnosed into three subtypes, having different disease course and severity stages: Relapsing Remitting (RR), Secondary Progressive (SP), and Primary Progressive (PP). Correct MS diagnosis, as well as subgroup identification and progression prediction, are of crucial relevance for the clinicians in order to provide the patient with personalized and efficient therapies. In addition clinically isolated syndrome (CIS) is a central nervous system demyelinating event isolated in time that is compatible with the possible future development of multiple sclerosis (MS). Early risk stratification for conversion of CIS to MS helps with treatment decisions. These tasks were all investigated. In particular a multimodal data set that included MRI data from Diffusion Weighted Images (DWI) and Magnetization Transfer Ratio (MTR) images was used. Diffusion metrics were extracted from DWI images: Fractional Anisotropy (FA); Mean Diffusivity (MD); Radial Diffusivity (RD); Axial Diffusivity (AD). Region of Interest (ROI) features were also derived. ROI measures were obtained by averaging the voxel values in predetermined brain regions, resulting in a low-dimensional brain representation.
In another data set, the extracted MRI features were: relaxometry measurements: quantitative proton density (qPD), T2 (qT2) and T1 (qT1); diffusion-derived measurements, namely intra-neurite volume fraction (intra), intrinsic diffusivity (diff) and neurite orientation dispersion entropy from the Spherical Mean Technique; cortical thickness (CT), region-of-interest (ROI) volume (vol) and quantitative total sodium concentration (TSC). The weighted mean of relaxometry, diffusion and sodium features over a probabilistic lesion map and normal appearing white matter were also added, for a total of 693 features. This cohort consisted of a total of 123 subjects: 29 HC, 18 CIS, 63 relapsing-remitting MS (RRMS) and 13 secondary progressive MS (SPMS) patients with same disease duration.
Classification task: patients vs controls
For the Multiple Sclerosis study, whole brain high-dimensional features were initially considered, resulting in poor predictive performances. Therefore, ROI features were used for the subsequent analysis. Based on single modalities, good classification results were obtained, with the Random Forest being the best performing classifier. This suggests that non-linear interactions among ROI based features are informative to predict MS.
The next step was to perform a multi-modal analysis. At this stage, it was confirmed that linear classifiers are not suitable for this task. Therefore, Multiple Kernel Learning techniques based on Gaussian Kernels as well as feature concatenation strategies with a Random Forest classifier were investigated. The results showed that a combination of features is often beneficial to improve the predictive performance. This approach in combination with Multiple Kernel Learning techniques is suitable to classify single patients into the correct peer group as needed for clinical decision support.
Classification task: disease progression
Predicting the progression of Multiple Sclerosis is known to be one of the most challenging tasks for both clinicians and data analysts. As there is no uniform consensus on the clinical assessment of disease severity itself, it is non-trivial to define exact class labels for the prediction task. On the available cohort, state-of-the-art classification models showed limited predictive performance on the disease progression task when tested to predict evolvement of patients with CIS to MS. A further investigation is recommended after a larger number of scans has been acquired, investigating the potential of complex deep learning based techniques and modern computer vision approaches. However our recent open access article in Frontiers (Johnson et al, Frontiers in neurology 2021) demonstrates that metrics from SMT (a technique for microstructure characterization developed within the CDS-QUAMRI project) correlate strongly with disability scores in MS, providing a simple non-invasive predictive marker that we plan to use widely in clinical studies and trials.
Classification task: Multiple Sclerosis subtypes
It was also investigated on the first cohort whether unsupervised algorithms were able to successfully identify clusters of patients reflecting the MS subtypes. As for the classification task, ROI features from diffusion images and MTR metrics were used as input. Various approaches were compared including DBSCAN, k-means, hierarchical clustering, and spectral clustering. When distinguishing between the three groups of MS (PP, SP, RR) a poor separation boundary was obtained. However, we observed that merging the groups of PP and SP patients helped to generally improve the results. In particular, both k-means and hierarchical clustering resulted in significant performances gains. As a final step, the potential to apply Principal Component Analysis (PCA) and cluster the subjects based on the first two components was investigated. This analysis resulted in a good separation of two groups of MS subtypes (SP + PP; RR) and can thus be used to classify single patients into the correct group as needed for clinical decision support. Furthermore, it was observed that only a subset of the ROI features was associated with the MS phenotype, suggesting that a lower dimensional representation would be beneficial. Given the sample size limitation, the number of clusters was considered as a fixed parameter. Future work should primarily focus on acquiring a larger data cohort. A sufficiently big sample size, improved hyperparameter optimization, and features combinations should be strived forto further enhance the clustering analysis.
On the second cohort a random forest classification over multiple tasks was performed: HC vs MS patients, HC vs CIS, CIS vs MS patients, and RRMS vs SPMS. Receiver operating characteristic (ROC) area under the curve (AUC) and Matthew’s correlation coefficient (MCC) were calculated to assess classification performances. Distinct features appeared to be relevant for each classification task. Within the top 15th percentile of variable importance, widespread qT2, volume and orientation dispersion entropy were the most relevant features for the classification of HC vs MS patients. Diffusion-derived microstructural features acquired more importance when CIS was introduced in the classification. Together with qT1, qT2 and volume, intra appeared to be important for CIS vs MS, while diff and TSC emerged when distinguishing HC vs CIS (Fig.2). A combination of features, including qT2, intra, volume and TSC characterised the RRMS vs SPMS classification. It appears that multi-modal qMRI provides unique microstructural features that are highly informative for distinguishing among MS subtypes and allow for correct classification of individua patients into the respective MS type. They convey complementary information to conventional macroscopic readouts, characterising disease heterogeneity at a level that may more closely reflect MS histopathology.
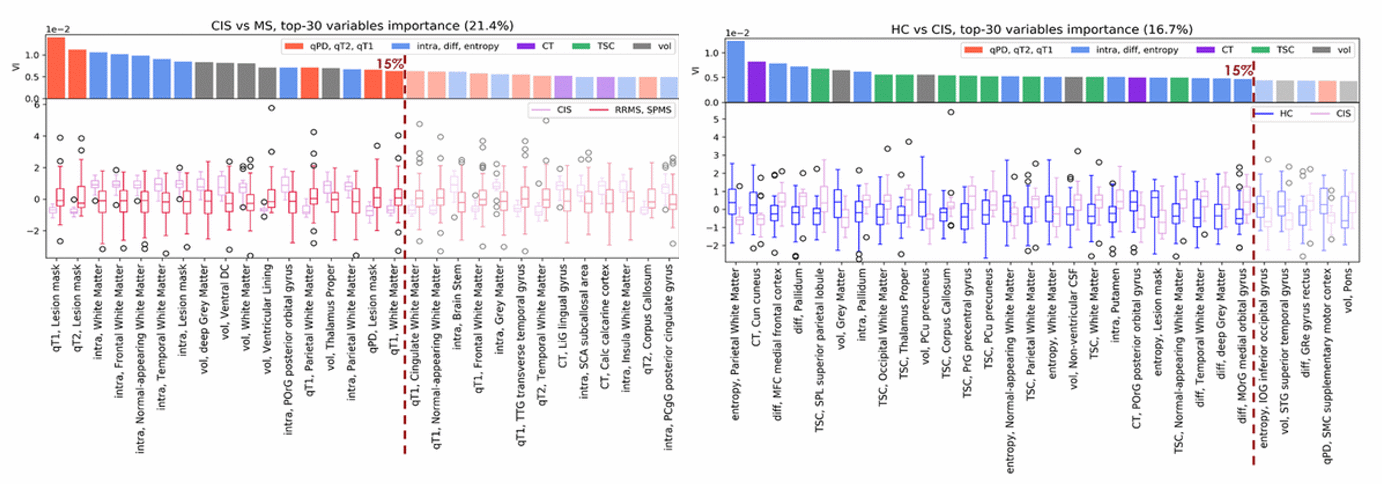
Figure 2. Contribution of CIS patients to the classification: CIS vs MS patients, HC vs CIS tasks. Inflammation in the probabilistic lesion map (higher qT2/qT1 in MS patients) and axonal loss (lower intra) in white matter (WM) were observed, together with atrophy (decreased tissue volume), in the classification of CIS vs MS patients. CIS showed also reduced microstructural integrity (mainly lower entropy in WM and higher diff in grey matter (GM) and deep GM) and higher TSC compared to HC.
